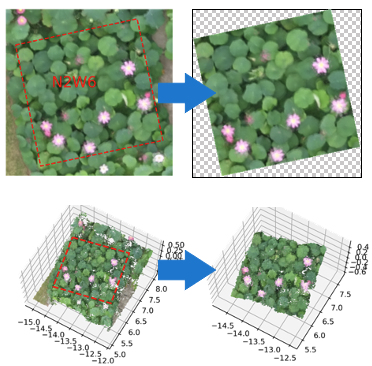
Welcome to EasyIDP’s documentation!#
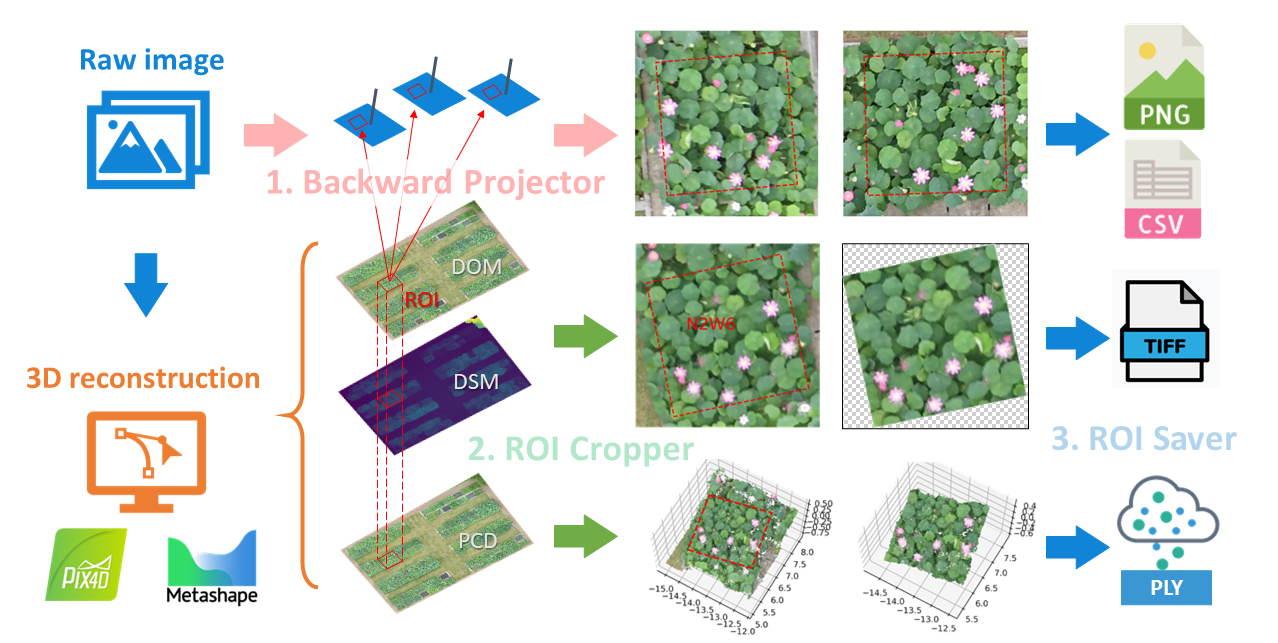
EasyIDP (Easy Intermediate Data Processor) is a handy tool for dealing with region of interest (ROI) on the image reconstruction (Metashape & Pix4D) outputs, mainly in agriculture applications. It provides the following functions:
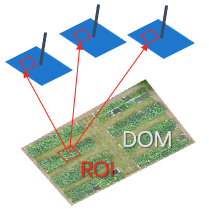
Backward Projection ROI to original images.
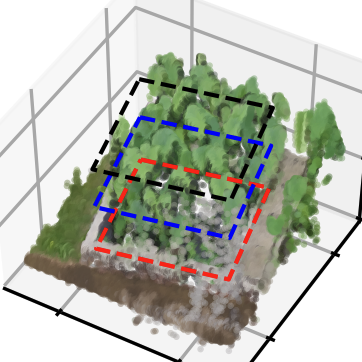
Clip ROI on GeoTiff Maps (DOM & DSM) and Point Cloud.
Save cropped results to corresponding files





other languages
This is a multi-language document, you can change document languages here or at the bottom left corner.
Note
In the EasyIDP, we use the (horizontal, vertical, dim) order as the coords order. When it applies to the GIS coordintes, is (longitude, latitude, altitude)
Examples#
Quick Start#
You can install the packages by PyPi:
pip install easyidp
Note
If you meet bugs in the pypi version, please consider using the latest source code. The tutorial can be found here: Using from source code.
And import the packages in your python code:
import easyidp as idp
Caution
Before doing the following example, please understand the basic pipeline for image 3D reconstruction by Pix4D or Metashape. And know how to export the DOM, DSM (*.tiff), and Point cloud (*.ply). Also require some basic knowledge about GIS shapefile format (*.shp).
1. Read ROI#
roi = idp.ROI("xxxx.shp") # lon and lat 2D info
# get z values from DSM
roi.get_z_from_dsm("xxxx_dsm.tiff") # add height 3D info
The 2D roi can be used to crop the DOM, DSM, and point cloud ( 2. Crop by ROI ). While the 3D roi can be used for Backward projection ( 4. Backward Projection )
Caution
It is highly recommended to ensure the shapefile and geotiff share the same coordinate reference systems (CRS), the built-in conversion algorithm in EasyIDP may suffer accuracy loss.
It is recommended to use Coordinate reference systems for “UTM” (WGS 84 / UTM grid system), the unit for x and y are in meters and have been tested by EasyIDP developers.
The traditional (longitude, latitude) coordinates like
epsg::4326also supported, but not recommended if you need calculate “distences” hence its unit is degree.The local country commonly used coordinates like BJZ54 (北京54), CGCS2000 (2000国家大地坐标系), JDG2011 (日本測地系2011), and etc., have not been tested and hard to guarantee workable in EasyIDP. Please convert to UTM by GIS software if you meet any problem.
The acceptable ROI types are only polygons (grids are a special type of polygon), and the size of each polygon should be fittable into the raw images (around the 1/4 size of one raw image should be the best).
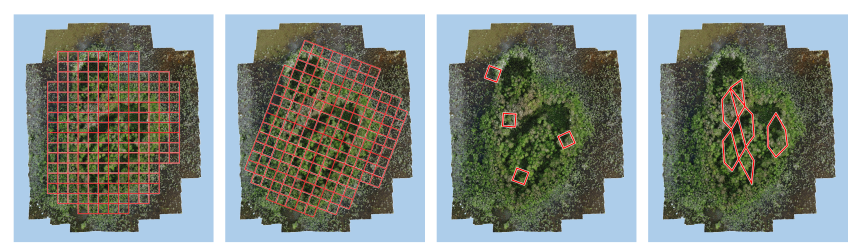
The fourth one may too large to be fitted into each raw image, recommend to make smaller polygons.#
2. Crop by ROI#
Read the DOM and DSM Geotiff Maps
dom = idp.GeoTiff("xxx_dom.tif")
dsm = idp.GeoTiff("xxx_dsm.tif")
Read point cloud data
ply = idp.PointCloud("xxx_pcd.ply")
crop the region of interest from ROI:
dom_parts = roi.crop(dom)
dsm_parts = roi.crop(dsm)
pcd_parts = roi.crop(ply)
If you want to save these crops to given folder:
dom_parts = roi.crop(dom, save_folder="./crop_dom")
dsm_parts = roi.crop(dsm, save_folder="./crop_dsm")
pcd_parts = roi.crop(ply, save_folder="./crop_pcd")
3. Read Reconstruction projects#
You can add the reconstructed plot individually or by batch adding
The Metashape projects naturally support using different chunks for different plots in one project file (*.psx), so the chunk_id is required to specify which plot are processing.
ms = idp.Metashape("xxxx.psx", chunk_id=0)
Caution
Though only the xxxx.psx is given here, the folder xxxx.files generated by Metashape is more important for EasyIDP. Please put them into the same directory.
Currently, the EasyIDP has not support parse the meta info that records the relative path to the raw image folder, so please manual specify the raw_img_folder if you need the backward projection.
p4d = idp.Pix4D(project_path="xxxx.p4d",
raw_img_folder="path/to/folders/with/raw/photos/",
# optional, in case you changed the pix4d project folder
param_folder="path/to/pix4d/parameter/folders")
Caution
Though only the xxxx.p4d is given here, the folder xxxx generated by Pix4D is more important for EasyIDP. Please put them into the same directory and not recommend the change the inner folder structure
4. Backward Projection#
img_dict = roi.back2raw(chunk1)
Then check the results:
# find the raw image name list
>>> img_dict.keys()
dict_keys(['DJI_0177', 'DJI_0178', 'DJI_0179', 'DJI_0180', ... ]
# the roi pixel coordinate on that image
>>> img_dict['DJI_0177']
array([[ 779, 902],
[1043, 846],
[1099, 1110],
[ 834, 1166],
[ 779, 902]])
Save backward projected images
img_dict = roi.back2raw(ms, save_folder="folder/to/put/results/")
img_dict = roi.back2raw(p4d, save_folder="folder/to/put/results/")
5. Forward Projection#
This function support bring results and image itself from corresponding raw images to geotiff.
Caution
The following features have not been supported.
Please using the following code to forward project from raw img to DOM (raw forward dom -> raw4dom):
This feather has not supported yet.
imarray_dict = roi.raw4dom(ms)
This is a dict contains the image ndarray of each ROI as keys, which projecting the part of raw image onto DOM.
>>> geotif = imarray_dict['roi_111']
>>> geotif
<class 'easyidp.geotiff.GeoTiff'>
You can also do the forward projecting with detected results, the polygon are in the geo coordinate. Before doing that, please prepare the detected results (by detection or segmentation, polygons in raw image pixel coordinates).
result_on_raw = idp.roi.load_results_on_raw_img(r"C:/path/to/detected/results/folder/IMG_2345.txt")
Then forward projecting by giving both to the function:
>>> imarray_dict, coord_dict = roi.raw4dom(ms, result_on_raw)
>>> coord_dict['roi_111']
{0: array([[139.54052962, 35.73475194],
...,
[139.54052962, 35.73475194]]),
...,
1: array([[139.54053488, 35.73473289],
...,
[139.54053488, 35.73473289]]),
Save single geotiff of one ROI:
>>> geotif = imarray_dict['roi_111']
>>> geotif.save(r"/file/path/to/save/geotif_roi_111.tif")
Batch save single geotiff of each ROI:
>>> imarray_dict = roi.raw4dom(ms, save_folder="/path/to/save/folder")
References#
Publication#
Please cite this paper if this project helps you:
@Article{wang_easyidp_2021,
AUTHOR = {Wang, Haozhou and Duan, Yulin and Shi, Yun and Kato, Yoichiro and Ninomiya, Seish and Guo, Wei},
TITLE = {EasyIDP: A Python Package for Intermediate Data Processing in UAV-Based Plant Phenotyping},
JOURNAL = {Remote Sensing},
VOLUME = {13},
YEAR = {2021},
NUMBER = {13},
ARTICLE-NUMBER = {2622},
URL = {https://www.mdpi.com/2072-4292/13/13/2622},
ISSN = {2072-4292},
DOI = {10.3390/rs13132622}
}
Site packages#
We also thanks the benefits from the following open source projects:
package main (for users)
numpy: https://numpy.org/
matplotlib: https://matplotlib.org/
scikit-image: https://github.com/scikit-image/scikit-image
tifffile: https://github.com/cgohlke/tifffile
imagecodecs: https://github.com/cgohlke/imagecodecs
shapely: https://github.com/shapely/shapely
laspy/lasrs/lasio: https://github.com/laspy/laspy
geojson: https://github.com/jazzband/geojson
package documentation (for developers)
nbsphinx: https://github.com/spatialaudio/nbsphinx
sphinx-gallery: https://github.com/sphinx-gallery/sphinx-gallery
sphinx-inline-tabs: https://github.com/pradyunsg/sphinx-inline-tabs
sphinx-intl: https://github.com/sphinx-doc/sphinx-intl
sphinx-rtc-theme: https://github.com/readthedocs/sphinx_rtd_theme
package testing and releasing (for developers)
packaging: https://github.com/pypa/packaging
Funding#
This project was partially funded by:
the JST AIP Acceleration Research “Studies of CPS platform to raise big-data-driven AI agriculture”;
the SICORP Program JPMJSC16H2;
CREST Programs JPMJCR16O2 and JPMJCR16O1;
the International Science & Technology Innovation Program of Chinese Academy of Agricultural Sciences (CAASTIP);
the National Natural Science Foundation of China U19A2